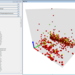
Visualization of viral evolution is one of the essential tasks in bioinformatics, through which virologists characterize a virus. The fundamental visualization tool for such a task is constructing a dendrogram, also called the phylogenetic tree. In this paper, we propose the visualization and characterization of the evolutionary path, starting from the root to isolated virus in the leaf of the phylogenetic tree. The suggested approach constructs the sequences of inner nodes (ancestors) within the phylogenetic tree and
uses one-hot-encoding to represent the genetic sequence in a binary format. By employing embedding methods, such as multi-dimensional scaling, we project the path into 2D and 3D spaces. The final visualization demonstrates the dynamic of viral evolution locally (for an individual strain) and globally (for all isolated viruses). The results suggest applications of our approach in: detecting earlier changes in the characteristics of strains; exploring emerging novel strains; modeling antigenic evolution; and study of evolution dynamics. All of these potential applications are critical in the fight against viruses.
Количество просмотров:
Вернуться в раздел Специализированные системы визуализации